Cas13 vs. Cas9: two different ways to potentially treat Angelman syndrome

by Ulrika Beitnere,
Segal Lab, UC Davis
There is a new kind of CRISPR nuclease on the gene technology block called Cas13. What is so special about it and what is the main difference between the better-known Cas9? This is a question not only many parents have been asking, but also scientists! You might want to know how Cas13 can be utilized for Angelman syndrome and why that strategy should be explored? "CRISPR gives us a new set of tools to use in the treatment of Angelman syndrome. The new Cas13 will allow us to target the antisense in a way that can be both long-term and highly specific,” states Dr. David Segal, UC Davis.
First, about the essential facts. Both Cas9 and Cas13 proteins are nucleases - enzymes that are able to cut phosphodiester bonds between nucleotides of nucleic acids. Both nucleases have been found to be programmable to almost any site in the genome. The main difference is that Cas13 targets and cuts RNA instead of DNA, and this could potentially result in less off-target effects. Since, in Angelman syndrome, the target is the long noncoding RNA known as the UBE3A-antisense transcript (Ube3a-ATS) avoiding a DNA target is optimal. DNA is the script for every cell in our body, whereas RNA is the working copy. Editing the working copy should be safer than mutating the original. With the understanding of RNA functions and critical roles in diseases, as well as the development of RNA-related technologies, there is a growing interest in developing novel RNA-based therapeutics, while avoiding a DNA target.
Recently, two independent studies used Cas9 to unsilence Ube3a by targeting it to a region on the Ube3a-ATS in Angelman syndrome in animal models. In the first study, Wolter et al. used an adeno-associated virus (AAV) to deliver Cas9 and found that the Cas9 caused the AAV to insert itself in the target region of the Ube3a-ATS in mice. Just a refresher - AAV is considered a small, safe virus that has been used as a delivery vehicle for two FDA-approved gene therapies already: Luxturna in 2017 for a rare inherited retinal dystrophy, and Zolgensma in 2019 for spinal muscular atrophy. For Wolter et al., the Cas9 caused the AAV to insert into the repetitive Snord115 cluster of genes located before the Ube3a gene (Figure 1A). By targeting a large number of target sites in that cluster, they were able to unsilence paternal Ube3a in neurons.
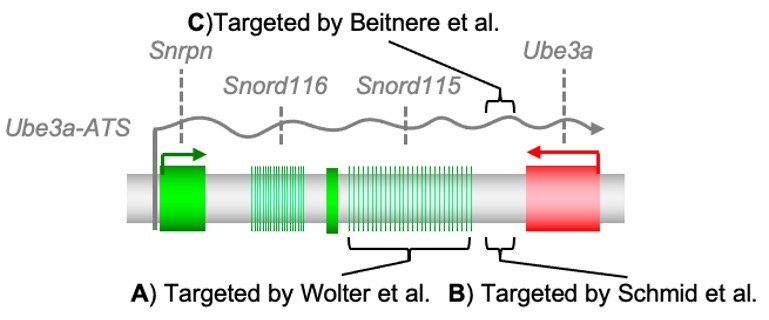
Figure 1. Targeting sites in the DNA (A, B) and RNA (C) forms of the Ube3a-ATS.
In the second study, Schmid et al. also used AAV for Cas9 delivery in a different site in the genome - between the Ube3a end which is called 3' UTR, and the Snord115 gene (Figure 1B). Unlike the Wolter et al. study, they did not see a lot of AAV insertion but instead the Cas9 introduced short insertions and deletions (indels) of the DNA at the target site. These indels interfere with the extension of the Ube3a-ATS and cause lasting expression of the Ube3a protein from the paternal allele. In summary, these two studies show the proof-of-principal for Cas9 to interfere with the Ube3a-ATS by creating double-stranded breaks in the DNA at different sites and with variable amounts of AAV integration can activate the paternal copy of the Ube3a gene and rescue AS behaviors in the AS mouse model. In the Segal lab, we are working to ultimately do the same job while targeting the RNA and not the DNA, in which there are no concerns about permanent changes to the DNA.
Cas13 is being used to target the Ube3a-ATS RNA without changing the DNA (Figure 1C). Using AAV to target Cas13 directly to the RNA form of the Ube3a-ATS in mice, we were also able to restore the expression of Ube3a from the paternal allele. Several studies are still underway so that we can publish our full results, but we are very excited about the potential of Cas13 to accomplish this task. Several other “RNA-targeted drugs” are in clinical trials, or approved, for medical use already: the antisense oligonucleotides (ASOs) used in the clinical trials for Angelman syndrome are a good example. Cas13 has the potential to be an ideal therapeutic for Angelman syndrome, just like ASOs, but without the need for repeated intrathecal injections every couple of months (Meng et al. 2015). For another RNA-targeted approach, RNA interference, three rare-disease treatments have already been approved by the FDA, including Onpattro (2018), Givlaari (2019), and Oxlumo (2020). When Cas13 was compared to RNA interference directly, Cas13 won in the precision department, revealing zero off-targets whereas the so-called short hairpin RNA showed numerous off-targets (Abudayyeh et al. 2017, Cox et al. 2017).
More exciting studies of Cas13 may be moving towards clinical trials. Early in 2020, when the COVID-19 virus was spreading like wildfire, a group of scientists showed that Cas13d can act as an antiviral treatment by successfully targeting for RNA knockdown of the Covid-19 and influenza A viruses (Abbott et al. 2020). Another showed that Cas13a can successfully be used for another virus disorder - hepatitis C (Ashraf et al. 2021). The first study using Cas13 in animals showed that Cas13 was able to knock down several genes in the liver using an AAV as the delivery vehicle (He et al. 2020).
In summary, the newer Cas13 class has the specificity and lack of off-targets that are ideal for clinical translation. Together, with the emerging approaches of Cas9 DNA-mediated gene knockdown, a strategy targeting RNA for disruption with Cas13 can be advantageous for the potential treatment of Angelman syndrome and we are very excited to be working on this project.
References
Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T, Endy D, La Russa MF, Lewis DB, Qi LS. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020 May 14;181(4):865-876.e12. doi: 10.1016/j.cell.2020.04.020. Epub 2020 Apr 29. PMID: 32353252; PMCID: PMC7189862.
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017 Oct 12;550(7675):280-284. doi: 10.1038/nature24049. Epub 2017 Oct 4. PMID: 28976959; PMCID: PMC5706658.
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017 Nov 24;358(6366):1019-1027. doi: 10.1126/science.aaq0180. Epub 2017 Oct 25. PMID: 29070703; PMCID: PMC5793859.
Ashraf MU, Salman HM, Khalid MF, Khan MHF, Anwar S, Afzal S, Idrees M, Chaudhary SU. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomed Pharmacother. 2021 Jan 19;136:111239. doi: 10.1016/j.biopha.2021.111239. Epub ahead of print. PMID: 33454599.
He B, Peng W, Huang J, Zhang H, Zhou Y, Yang X, Liu J, Li Z, Xu C, Xue M, Yang H, Huang P. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell. 2020 Jul;11(7):518-524. doi: 10.1007/s13238-020-00700-2. PMID: 32185621; PMCID: PMC7095259.
Hebras J, Marty V, Personnaz J, Mercier P, Krogh N, Nielsen H, Aguirrebengoa M, Seitz H, Pradere JP, Guiard BP, Cavaille J. Reassessment of the involvement of Snord115 in the serotonin 2c receptor pathway in a genetically relevant mouse model. Elife. 2020 Oct 5;9:e60862. doi: 10.7554/eLife.60862. PMID: 33016258; PMCID: PMC7673782.
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018 Apr 19;173(3):665-676.e14. doi: 10.1016/j.cell.2018.02.033. Epub 2018 Mar 15. PMID: 29551272; PMCID: PMC5910255.
Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015 Feb 19;518(7539):409-12. doi: 10.1038/nature13975. Epub 2014 Dec 1. PMID: 25470045; PMCID: PMC4351819.