Part I: Introducing CRISPR, A Promising Gene Editing Technology
Barb Bailus, PhD
David Gurzick, PhD
Hello Everyone,
Over the next few weeks, we are going to be looking at the technology behind many of the gene therapy techniques that are showing tremendous potential for application in Angelman syndrome. We’ll start with some of the background science and then dive in to talk about how they are being used by our team of researchers.
It seems the perfect place to start is with CRISPR – the technology that has garnered the most public attention as of late.
CRISPR, short for Clustered Regularly Interspaced Short Palindromic Repeats, arrived on the gene editing scene back in 2013. This has quickly taken over the gene editing field. It is now in use in 1000s of labs around the world and developments are coming out daily.
Back in 2013, scientists Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier were among the first to harness the amazing gene editing power of CRISPR. CRISPR originated in bacteria, being used as an immune system. When bacteria get a viral infection, the CRISPR system fights off the virus by making a guide RNA (or gRNA for short). Think of gRNA as an address label to the viral DNA, which CRISPR uses to target the viral DNA and cut it into pieces. CRISPR then saves a piece of that viral DNA so the next time the bacteria is infected by the same virus it fights it off even faster. Scientists have been able to modify the CRISPR system so that it can actually work in humans, plants, and other animals in order to modify the DNA of the desired organism.
This CRISPR system is what is being used to cut and replace DNA in gene editing. In scientific use, the term CRISPR is always followed by the name of a protein. Cas9 (pronounced “kass nine”) is the most common and was among the first to be used successfully. Cas9, like other proteins, uses a piece of gRNA to figure out where to cut our DNA. Imagine you have a protein like UBE3A that we know is incorrectly configured. You create a guide RNA that matches the incorrect pattern and the Cas9 will hunt for it and when it finds a match, it will snip it out. Don’t scream “Yeah! No more incorrect UBE3A!” just yet.There is also the need to replace what was cut out; otherwise your DNA would remain damaged in this area. This occurs through the cell’s natural process of repair in a fashion similar to what a parent does when their child spits out their pacifier – the enzymes of the repair process essentially grab for whatever is immediately nearby that looks like a fit and puts it in.
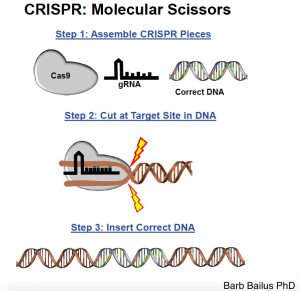 In practice this works by injecting both a large number of vectors with the gRNA – Cas9 in most cases – and a bunch of desired sequences of genome. So Cas9 cuts out the part you want it to and inserts the desired sequence. Gene editing is then complete.
CRISPR Cas9 can also be used to cut the antisense transcript on the paternal allele of of chromosome 15, unsilencing the normal silent copy. This is an approach that many scientists are currently exploring in Angelman syndrome This will be further discussed in upcoming blogs on CRISPR.
This is certainly an oversimplification and there are lots of caveats that the field is working through. One example is off-target edits, where the process clips out wrong places that still match the exact, or a similar, pattern of the gRNA. This can be potentially dangerous, and is being carefully worked out and incrementally modified by many pioneering scientists. The NIH is aggressively backing this type of research as well. In June of 2019, they awarded UC Davis $9m to create a center to advance genome-editing tools and to serve as the nation’s testing site for safety and efficacy. FAST researcher David Segal is among the leadership team announced for this institute.
Oh, and by the way, CRISPR is not limited to humans. The process can be used in plant and animal systems. Indeed, this is the process that FAST scientists have used to create mice, rats, and other animal models of Angelman syndrome for testing; editing them to contain defective, or missing, copies of UBE3A.
CRISPR is not the only game in town. There are other technologies that support gene editing, including a process called zinc fingers (see footnote). CRISPR is special, however, due to its ease of use and versatility; suddenly a laboratory that specialized in a specific disease didn’t have to hire a genetic engineering expert. Instead, the laboratory could buy a “CRISPR Kit” and edit the genome to their desired specifications. This ease of editing has had a huge impact on our understanding of human disease, creating better models and potential disease therapies.
The beauty of this system is in both its simplicity and flexibility. With CRISPR, the Cas9 is always the same tool being used for editing; a scientist doesn’t have to build and test a new tool for each target gene, unlike zinc fingers, which have to be re-designed for every target gene. The gRNA can be ordered and synthesized from a company, making it very easy and fast for scientists to use. Additionally, having the Cas9 be the same tool for all gene targeting allows for a researcher to target multiple gene targets simultaneously by just providing multiple gRNA address labels; a revolutionary step forward for multigene disease (Alzheimer’s, Parkinson’s, Autism etc).
In an upcoming blog entry, we are going to explore how this amazing tool can be used to further our understanding of human disease, and the potential ways to use CRISPR for different gene editing therapies. Until then here is an excellent video on CRISPR.
---
Footnote
[1] During the 1990s, scientists at public universities and a private company, Celera, sequenced the first human genome. This initial map of the human genome was essential in identifying disease-associated genes. Soon after the human genome was sequenced scientists began to search for ways that the genome could be modified, either by directly modifying the genetic sequence, or by modifying gene expression. Zinc fingers were one of first widely used tools for gene engineering. Zinc fingers required extensive expertise to use though, limiting the laboratories that could employ this technology. One of the advantages of zinc finger proteins is that they are found in mammals, and are therefore unlikely to elicit an immune response when used to treat human patients. This technology is the basis for a large company, Sangamo, which has brought zinc fingers into human clinical trials for various diseases. In 2012, a new gene editing enzyme, CRISPR, was characterized and modified for editing a specific DNA sequence in the labs of Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier.
In practice this works by injecting both a large number of vectors with the gRNA – Cas9 in most cases – and a bunch of desired sequences of genome. So Cas9 cuts out the part you want it to and inserts the desired sequence. Gene editing is then complete.
CRISPR Cas9 can also be used to cut the antisense transcript on the paternal allele of of chromosome 15, unsilencing the normal silent copy. This is an approach that many scientists are currently exploring in Angelman syndrome This will be further discussed in upcoming blogs on CRISPR.
This is certainly an oversimplification and there are lots of caveats that the field is working through. One example is off-target edits, where the process clips out wrong places that still match the exact, or a similar, pattern of the gRNA. This can be potentially dangerous, and is being carefully worked out and incrementally modified by many pioneering scientists. The NIH is aggressively backing this type of research as well. In June of 2019, they awarded UC Davis $9m to create a center to advance genome-editing tools and to serve as the nation’s testing site for safety and efficacy. FAST researcher David Segal is among the leadership team announced for this institute.
Oh, and by the way, CRISPR is not limited to humans. The process can be used in plant and animal systems. Indeed, this is the process that FAST scientists have used to create mice, rats, and other animal models of Angelman syndrome for testing; editing them to contain defective, or missing, copies of UBE3A.
CRISPR is not the only game in town. There are other technologies that support gene editing, including a process called zinc fingers (see footnote). CRISPR is special, however, due to its ease of use and versatility; suddenly a laboratory that specialized in a specific disease didn’t have to hire a genetic engineering expert. Instead, the laboratory could buy a “CRISPR Kit” and edit the genome to their desired specifications. This ease of editing has had a huge impact on our understanding of human disease, creating better models and potential disease therapies.
The beauty of this system is in both its simplicity and flexibility. With CRISPR, the Cas9 is always the same tool being used for editing; a scientist doesn’t have to build and test a new tool for each target gene, unlike zinc fingers, which have to be re-designed for every target gene. The gRNA can be ordered and synthesized from a company, making it very easy and fast for scientists to use. Additionally, having the Cas9 be the same tool for all gene targeting allows for a researcher to target multiple gene targets simultaneously by just providing multiple gRNA address labels; a revolutionary step forward for multigene disease (Alzheimer’s, Parkinson’s, Autism etc).
In an upcoming blog entry, we are going to explore how this amazing tool can be used to further our understanding of human disease, and the potential ways to use CRISPR for different gene editing therapies. Until then here is an excellent video on CRISPR.
---
Footnote
[1] During the 1990s, scientists at public universities and a private company, Celera, sequenced the first human genome. This initial map of the human genome was essential in identifying disease-associated genes. Soon after the human genome was sequenced scientists began to search for ways that the genome could be modified, either by directly modifying the genetic sequence, or by modifying gene expression. Zinc fingers were one of first widely used tools for gene engineering. Zinc fingers required extensive expertise to use though, limiting the laboratories that could employ this technology. One of the advantages of zinc finger proteins is that they are found in mammals, and are therefore unlikely to elicit an immune response when used to treat human patients. This technology is the basis for a large company, Sangamo, which has brought zinc fingers into human clinical trials for various diseases. In 2012, a new gene editing enzyme, CRISPR, was characterized and modified for editing a specific DNA sequence in the labs of Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier.
 In practice this works by injecting both a large number of vectors with the gRNA – Cas9 in most cases – and a bunch of desired sequences of genome. So Cas9 cuts out the part you want it to and inserts the desired sequence. Gene editing is then complete.
CRISPR Cas9 can also be used to cut the antisense transcript on the paternal allele of of chromosome 15, unsilencing the normal silent copy. This is an approach that many scientists are currently exploring in Angelman syndrome This will be further discussed in upcoming blogs on CRISPR.
This is certainly an oversimplification and there are lots of caveats that the field is working through. One example is off-target edits, where the process clips out wrong places that still match the exact, or a similar, pattern of the gRNA. This can be potentially dangerous, and is being carefully worked out and incrementally modified by many pioneering scientists. The NIH is aggressively backing this type of research as well. In June of 2019, they awarded UC Davis $9m to create a center to advance genome-editing tools and to serve as the nation’s testing site for safety and efficacy. FAST researcher David Segal is among the leadership team announced for this institute.
Oh, and by the way, CRISPR is not limited to humans. The process can be used in plant and animal systems. Indeed, this is the process that FAST scientists have used to create mice, rats, and other animal models of Angelman syndrome for testing; editing them to contain defective, or missing, copies of UBE3A.
CRISPR is not the only game in town. There are other technologies that support gene editing, including a process called zinc fingers (see footnote). CRISPR is special, however, due to its ease of use and versatility; suddenly a laboratory that specialized in a specific disease didn’t have to hire a genetic engineering expert. Instead, the laboratory could buy a “CRISPR Kit” and edit the genome to their desired specifications. This ease of editing has had a huge impact on our understanding of human disease, creating better models and potential disease therapies.
The beauty of this system is in both its simplicity and flexibility. With CRISPR, the Cas9 is always the same tool being used for editing; a scientist doesn’t have to build and test a new tool for each target gene, unlike zinc fingers, which have to be re-designed for every target gene. The gRNA can be ordered and synthesized from a company, making it very easy and fast for scientists to use. Additionally, having the Cas9 be the same tool for all gene targeting allows for a researcher to target multiple gene targets simultaneously by just providing multiple gRNA address labels; a revolutionary step forward for multigene disease (Alzheimer’s, Parkinson’s, Autism etc).
In an upcoming blog entry, we are going to explore how this amazing tool can be used to further our understanding of human disease, and the potential ways to use CRISPR for different gene editing therapies. Until then here is an excellent video on CRISPR.
---
Footnote
[1] During the 1990s, scientists at public universities and a private company, Celera, sequenced the first human genome. This initial map of the human genome was essential in identifying disease-associated genes. Soon after the human genome was sequenced scientists began to search for ways that the genome could be modified, either by directly modifying the genetic sequence, or by modifying gene expression. Zinc fingers were one of first widely used tools for gene engineering. Zinc fingers required extensive expertise to use though, limiting the laboratories that could employ this technology. One of the advantages of zinc finger proteins is that they are found in mammals, and are therefore unlikely to elicit an immune response when used to treat human patients. This technology is the basis for a large company, Sangamo, which has brought zinc fingers into human clinical trials for various diseases. In 2012, a new gene editing enzyme, CRISPR, was characterized and modified for editing a specific DNA sequence in the labs of Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier.
In practice this works by injecting both a large number of vectors with the gRNA – Cas9 in most cases – and a bunch of desired sequences of genome. So Cas9 cuts out the part you want it to and inserts the desired sequence. Gene editing is then complete.
CRISPR Cas9 can also be used to cut the antisense transcript on the paternal allele of of chromosome 15, unsilencing the normal silent copy. This is an approach that many scientists are currently exploring in Angelman syndrome This will be further discussed in upcoming blogs on CRISPR.
This is certainly an oversimplification and there are lots of caveats that the field is working through. One example is off-target edits, where the process clips out wrong places that still match the exact, or a similar, pattern of the gRNA. This can be potentially dangerous, and is being carefully worked out and incrementally modified by many pioneering scientists. The NIH is aggressively backing this type of research as well. In June of 2019, they awarded UC Davis $9m to create a center to advance genome-editing tools and to serve as the nation’s testing site for safety and efficacy. FAST researcher David Segal is among the leadership team announced for this institute.
Oh, and by the way, CRISPR is not limited to humans. The process can be used in plant and animal systems. Indeed, this is the process that FAST scientists have used to create mice, rats, and other animal models of Angelman syndrome for testing; editing them to contain defective, or missing, copies of UBE3A.
CRISPR is not the only game in town. There are other technologies that support gene editing, including a process called zinc fingers (see footnote). CRISPR is special, however, due to its ease of use and versatility; suddenly a laboratory that specialized in a specific disease didn’t have to hire a genetic engineering expert. Instead, the laboratory could buy a “CRISPR Kit” and edit the genome to their desired specifications. This ease of editing has had a huge impact on our understanding of human disease, creating better models and potential disease therapies.
The beauty of this system is in both its simplicity and flexibility. With CRISPR, the Cas9 is always the same tool being used for editing; a scientist doesn’t have to build and test a new tool for each target gene, unlike zinc fingers, which have to be re-designed for every target gene. The gRNA can be ordered and synthesized from a company, making it very easy and fast for scientists to use. Additionally, having the Cas9 be the same tool for all gene targeting allows for a researcher to target multiple gene targets simultaneously by just providing multiple gRNA address labels; a revolutionary step forward for multigene disease (Alzheimer’s, Parkinson’s, Autism etc).
In an upcoming blog entry, we are going to explore how this amazing tool can be used to further our understanding of human disease, and the potential ways to use CRISPR for different gene editing therapies. Until then here is an excellent video on CRISPR.
---
Footnote
[1] During the 1990s, scientists at public universities and a private company, Celera, sequenced the first human genome. This initial map of the human genome was essential in identifying disease-associated genes. Soon after the human genome was sequenced scientists began to search for ways that the genome could be modified, either by directly modifying the genetic sequence, or by modifying gene expression. Zinc fingers were one of first widely used tools for gene engineering. Zinc fingers required extensive expertise to use though, limiting the laboratories that could employ this technology. One of the advantages of zinc finger proteins is that they are found in mammals, and are therefore unlikely to elicit an immune response when used to treat human patients. This technology is the basis for a large company, Sangamo, which has brought zinc fingers into human clinical trials for various diseases. In 2012, a new gene editing enzyme, CRISPR, was characterized and modified for editing a specific DNA sequence in the labs of Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier.